Goslin webapplication - A Parser, Validator and Normalizer for Shorthand Lipid Nomenclature
jGoslin is a parser implementation for lipid species and subspecies names following the most recent updates to the lipid shorthand nomenclature, the previous shorthand notation for lipids , the LIPID MAPS® Nomenclature (for shorthand names), the Swiss Lipids nomenclature and some more. We collaborate with the Lipidomics Standards Initiative concerning lipid nomenclature used for reporting of lipid species identifications.
The parser implementation is based on the Goslin grammars and returns a classification of the parsed lipid name including its perceived level (e.g. species level), including fatty acid and long chain base details, if they are deducable from the provided string. The hierarchy levels and the names are consistent with those used in the recent updates to the lipid shorthand nomenclature.
This web application parses the submitted lipid shorthand names and translates them into a normalized hierarchical representation either via the two submission forms below or via the REST API.
Please check the further information and specifically the Lipid Classes table to find out, whether a particular lipid class is already supported. Also, if you find errors or inconsistencies, please do not hesitate to either file a bug at GitHub, or to contact us.
LIFS is a project funded by BMBF as part of the German network for bioinformatics infrastructure (de.NBI) to provide and support tools for lipid-related bioinformatics.

News and Updates
-
This release adds an updated funding logo, replacing the BMBF logo with the new BMFTR one. To track progress of Goslin updates, please check our jgoslin release page and GitHub issues page.
-
This release adds mappings of lipid names to ChEbi entries (version as of Jan 18th, 2024). Please note that the user interface will now return original names of matched lipid shorthand names in other databases, not the shorthand name. The REST response and tabular output will contain direct URLs to the corresponding entries in the external databases. To track progress of Goslin updates, please check our jgoslin release page and GitHub issues page.
-
This release updates the internal implementation to use Goslin 2.2.0. This release improves support for mediators, gangliosides, HMDB name support and parsing of adducts with labeled isotopes. It also fixes wrong lyso classifications for SPB, SPBP, LHexCer and LSM. The web application now outputs the names of each lipid on all supported levels of the shorthand name hierarchy, both for tabular file export and REST API. To track progress of Goslin updates, please check our jgoslin release page and GitHub issues page.
-
This release fixes the REST API description. To track progress of Goslin updates, please check our jgoslin release page and GitHub issues page.
-
This release updates the internal implementation to use Goslin 2.1.0 fixing various issues and adding support for more lipid classes, deuterated lipids and improved support for Glycosphingolipids. This update includes the updated LIPID MAPS LMSD database as of March 3rd 2023. To track progress of Goslin updates, please check our jgoslin release page and GitHub issues page.
-
This release updates the internal implementation to use Goslin 2.0.1 to fix an issue with normalized head group names. To track progress of Goslin updates, please check our GitHub issues page.
-
This release updates the internal implementation to use Goslin 2.0, which supports the updated LIPID MAPS nomenclature and shorthand notation. We have also updated the LIPID MAPS database mapping to the latest version as of October 28th, 2021. To track progress of Goslin updates, please check our GitHub issues page.
-
This release fixes some issues with database loading and properties introduced by an update of the Spring Boot library.
To track progress of Goslin Updates please check our GitHub issues page.
-
This release updates the matomo tracking url. To track progress of Goslin updates, please check our GitHub issues page.
-
This release updates the system libraries to their latest versions and completes the migration to Keycloak. Please note that this release does not yet support the updated LIPID MAPS nomenclature and shorthand notation. To track progress of Goslin updates, please check our GitHub issues page.
-
This release updates the LIPID MAPS database mapping to the latest version as of October 27th, 2020. Please note that this release does not yet support the updated LIPID MAPS nomenclature and shorthand notation. To track progress of Goslin updates, please check our GitHub issues page.
-
This release follows release 1.1.2 of the jgoslin library. It adds additional warnings whenever modifications are encountered. At the moment, only hydroxylations are properly accounted for within the mass and sum formula calculation. It also updates the jgoslin dependency to the latest release version 1.1.2.
-
This release adds support for mass and sum formula calculation for most lipid classes (please check the lipid class table) based on the neutral lipid. It also adds support for adducts within the Goslin and Goslin fragments grammars, currently limited to those supported by LipidCreator. Thus, normalized lipid shorthand notation names, as provided after translation with Goslin, can now be combined with an adduct. Please note that adduct charges need to be explicit, thus '[M+H]+' needs to be provided as '[M+H]1+'. Adducts are also factored into the mass and sum formula calculations.
This release contains mappings for normalized lipid names to SwissLipids (updated May 15th, 2020) and LipidMaps (updated February 10th, 2020). The Goslin web application is not a meta search engine (yet).
-
Initial public release version of the Goslin web application. This release contains support for automatic linking of normalized lipid names to SwissLipids and LIPID MAPS® entries. This requires preprocessing of a subset of the LIPID MAPS® structure database (LMSD) and the SwissLipids 'lipids' database. Each lipid name and abbreviations thereof, if available, are parsed with the parser specific to the database and normalized. This normalized lipid name is the used as a dynamic lookup by the goslin web application to retrieve the matching entry URIs from the preprocessed database tables.
Lipid Shorthand Nomenclature Grammars
Goslin uses ANTLRv4 compatible context-free EBNF grammars. All Goslin implementations use a generic recursive decent parser (see Context-free language, LL parser, ANTLRv4).
These grammars (*.g4 files) are available from our Goslin GitHub repository at https://github.com/lifs-tools/goslin.
Hierarchical Representation of Lipids
The grammars model lipids as hierarchically structured bits of information.
We do not model the lipid category or main class explicitly, but rather keep them in a global lookup table data structure, derived from the lipid-list.csv file for reference.
This allows us to keep the grammars clutter-free and makes them easier to read.
The structural classification of lipids follows the shorthand notation recently updated by Liebisch et al. and is compatible to that of LIPID MAPS. The following example shows the hierarchical representation of PE 16:1(6Z)/16:0;5OH[R],8OH;3oxo:
Table 1. Structural hierarchy representation of PE 16:1(6Z)/16:0;5OH[R],8OH;3oxo). LM: LIPID MAPS, HG: Head Group, FA: Fatty AcylLevel |
Name |
Description |
Category (LM) |
Glycerophospholipids (GP) |
Lipid category |
Class (LM) |
Glycerophosphoethanolamine (PE) GP02 |
Lipid class |
Species (LM Subclass) |
Phosphatidylethanolamine, PE 32:2;O3 |
HG, FA summary, two double bond equivalents, three oxidations |
Molecular species |
PE 16:1_16:1;O3 |
HG, two FAs, two double bond equivalents, three oxidations |
sn-Position |
PE 16:1/16:1;O3 |
HG, SN positions, here: for FA1 at sn1 and FA2 at sn2, two double bond equivalents, three oxidations |
Structure defined |
PE 16:1(6)/16:1;(OH)2;oxo |
HG, SN positions, here: for FA1 at sn1 and FA2 at sn2, three oxidations and unspecified stereo configuration (6) on FA1 |
Full structure |
PE 16:1(6Z)/16:1;5OH,8OH;3oxo |
HG, SN positions, here: for FA1 at sn1 and FA2 at sn2, positions for oxidations and stereo configuration (6Z) on FA1 |
Complete structure |
PE 16:1(6Z)/16:0;5OH[R],8OH;3oxo |
HG, SN positions, here: for FA1 at sn1 and FA2 at sn2, positions for oxidations and stereo configuration ([R]) and double bond position and stereo configuration (6Z) on FA1 |
Please see the Goslin object model for an overview of the Goslin domain model which is used to represent the structural hierarchy by the different implementations.
Web Application and REST API
Interactive Usage
The interactive goslin web application is available
at https://apps.lifs-tools.org/goslin. It provides two forms to i) upload
a file containing one lipid name per line (see Figure 1.), or ii)
upload a list of lipid names, defined by the user in an interactive form
(see Figure 2). The
latter form also allows pasting lists of lipid names directly from the
clipboard with CTRL+V. Both forms provide feedback for issues
concerning every processed lipid, such as invalid names or typos (see Figure 3.), to
allow the user to cross-check their data before proceeding.
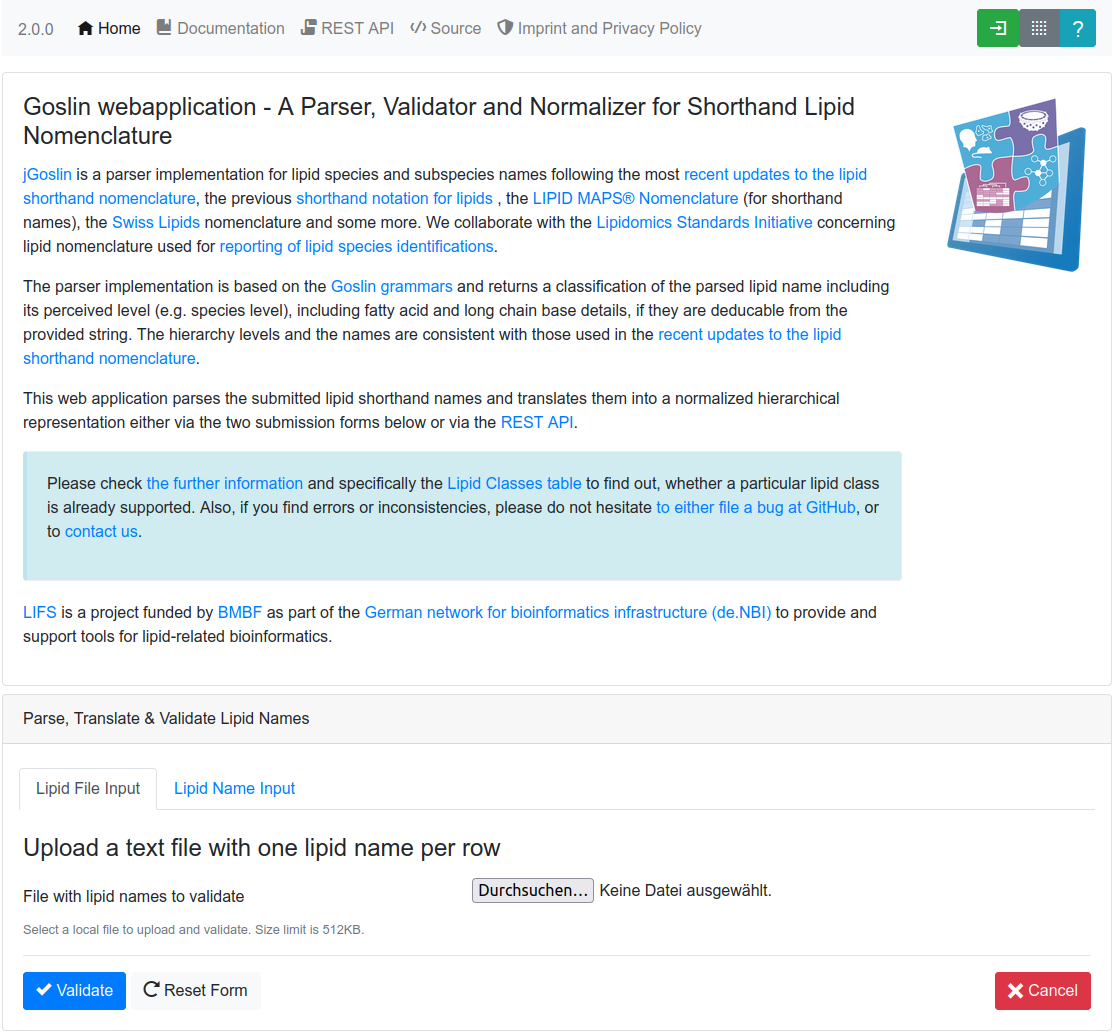

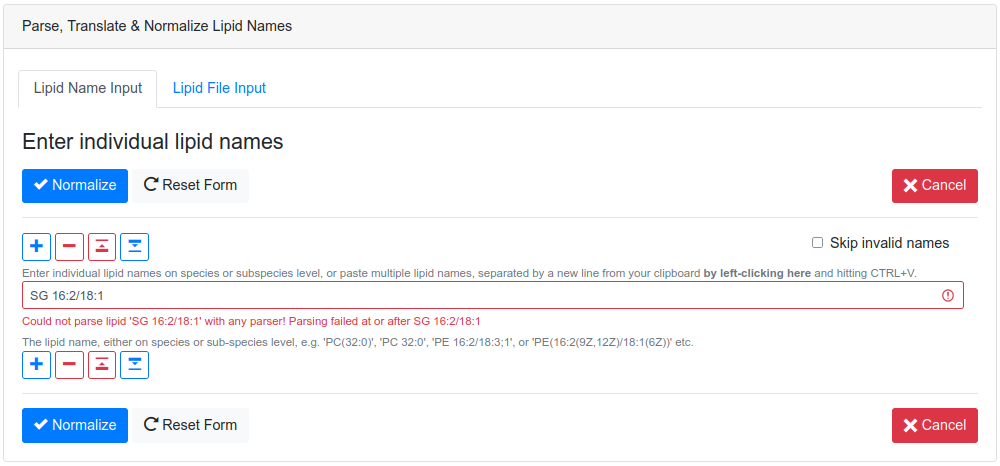
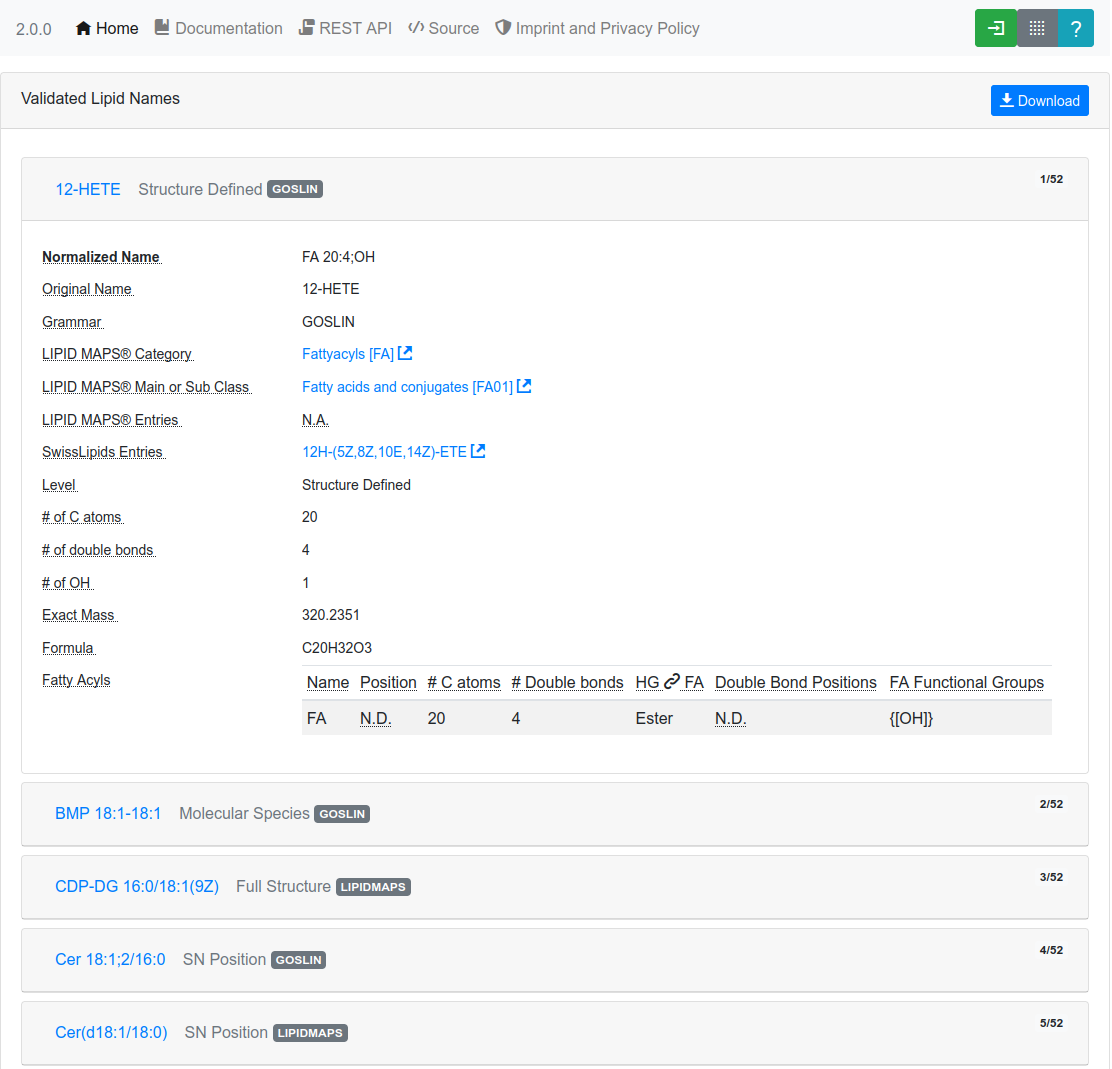
After successful validation, the validated lipids are returned in overview cards (see Figure 4.), detailing their LIPID MAPS® classification, cross-links to SwissLipids and/or LIPID MAPS®. Additionally, the cards show summary information about the number of carbon atoms, double bonds, hydroxylations and detailed information, such as double bond position, long-chain-base status, and the bond type of the fatty acyl to the head group for each fatty acyl, if available (see Figure 5.).
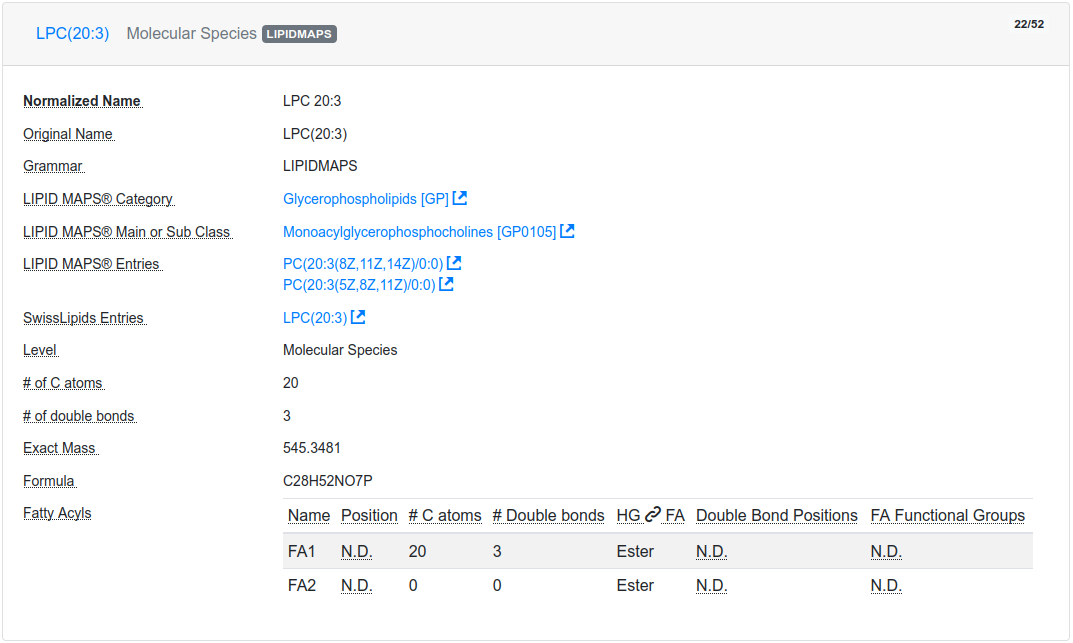
Finally, it is also possible to download the translated list of lipid names and their properties as a tab-separated file, by clicking on the Download button. Depending on the lipid level, undefined sn positions of regular lipids will be reported as "-1" in the table and as "0" for solitary fatty acids.
The source code for the web application and instructions to build it as a Docker container are available at https://github.com/lifs-tools/goslin-webapp under the terms of the open source Apache license version 2.
Programmatic access via the REST API
An interactive documentation for the rest api of the goslin web application is available at https://apps.lifs-tools.org/goslin/swagger-ui.html (see Figure 6.). To illustrate its usage, we will briefly show a small example how a user can access the rest api with a standard http client.
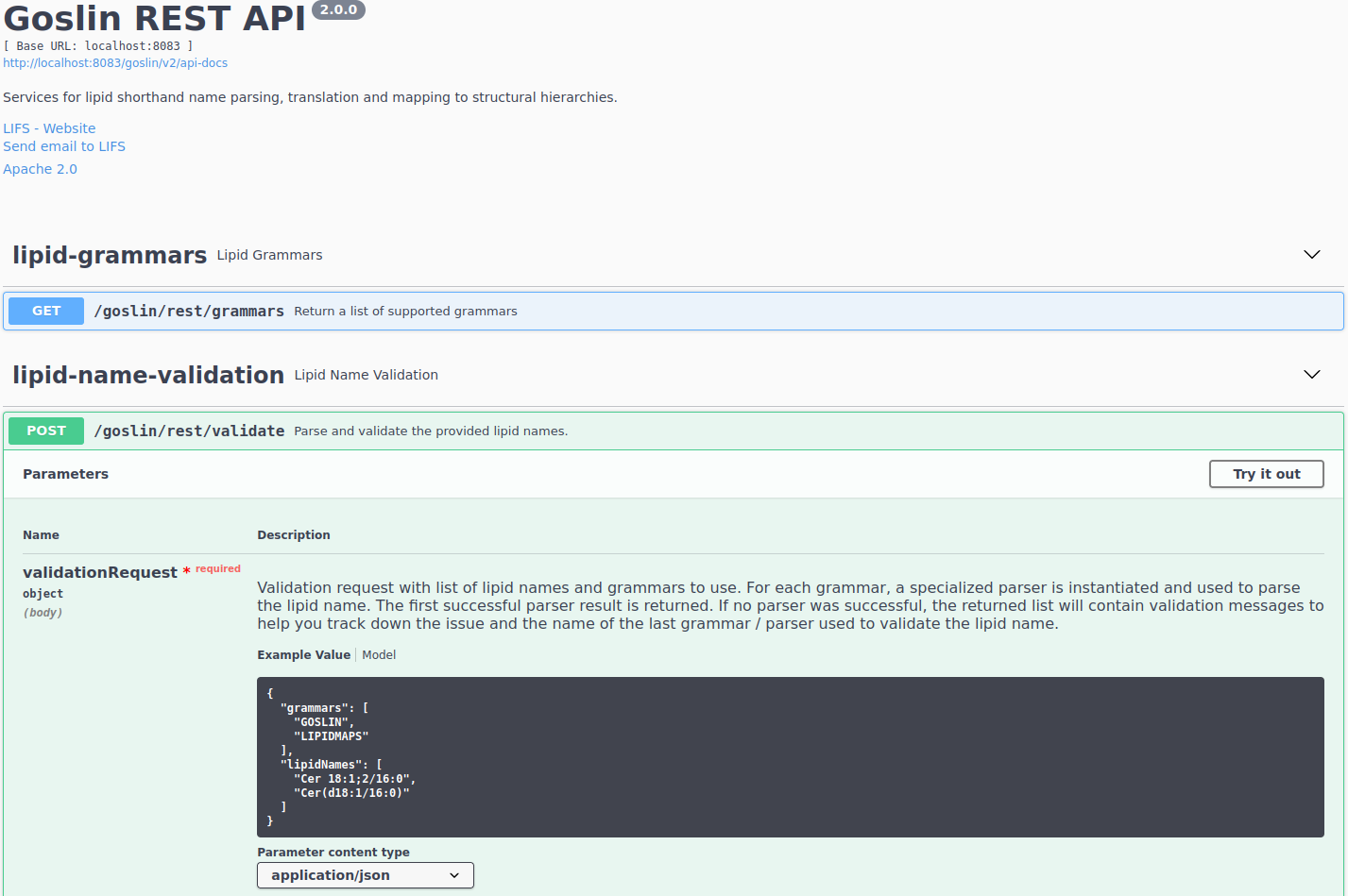
The Structure for the request consists of a json object \{} enclosing
two lists, with the names lipidNames and grammars. Acceptable values
for grammars are: LIPIDMAPS, GOSLIN, SHORTHAND, FATTY_ACID,
SWISSLIPIDS, and HMDB. A complete list is available from the
interactive rest api documentation’s Models section under
ValidationRequest. The available grammars may also be retrieved by the grammars method. Both fields in the ValidationRequest accept
comma-separated entries, enclosed in double quotes:
{
"lipidNames": [
"Cer(d18:1/16:1(6Z))"
],
"grammars": [
"LIPIDMAPS"
]
}
Sending the http POST request with curl as an http client looks as
follows:
curl -X POST "https://apps.lifs-tools.org/goslin/rest/validate" -H "accept: */*" -H "Content-Type: application/json" -d "{ \"lipidNames\": [ \"Cer(d18:1/16:1(6Z))\" ], \"grammars\": [ \"LIPIDMAPS\" ]}"
The rest api will return the following result for the request, with a
http response code of 200 (OK). This result returns a map of properties
for each lipid name that was parsed. If at least one name is not
parseable, the rest api will return a response code of 400 (Client
error), together with the same results reponse object. In that case, the
failedToParse field in the response will contain the number of lipid
names that could not be parsed. For those results where no grammar was
applicable, the grammar field will contain the string
NOT_PARSEABLE.¸In other cases, that field will contain the last
grammar used to parse the lipid name and the messages field will
contain a list of validation messages that help to narrow down the
offending bits in the lipid name.
{
"results": [
{
"lipidName": "Cer(d18:1/16:1(6Z))",
"grammar": "LIPIDMAPS",
"messages": [],
"lipidAdduct": {
"lipid": {
"headGroup": {
"headgroup": "Cer",
"lipidCategory": "SP",
"lipidClass": 29,
"className": "Cer"
},
"info": {
"name": "info",
"position": 0,
"count": 1,
"doubleBonds": {
"numDoubleBonds": 2,
"doubleBondPositions": {}
},
"functionalGroups": {
"OH": [
{
"name": "OH",
"position": -1,
"count": 2,
"doubleBonds": {
"numDoubleBonds": 0,
"doubleBondPositions": {}
},
"functionalGroups": {},
"ndoubleBonds": 0
}
]
},
"numCarbon": 34,
"lipidFaBondType": "LCB_EXCEPTION",
"level": "SN_POSITION",
"numEthers": 0,
"numSpecifiedFa": 1,
"totalFa": 2,
"extendedClass": "ESTER",
"lipidClass": 29,
"ndoubleBonds": 2
},
"faList": [
{
"name": "LCB",
"position": 1,
"count": 1,
"doubleBonds": {
"numDoubleBonds": 1,
"doubleBondPositions": {}
},
"functionalGroups": {
"OH": [
{
"name": "OH",
"position": -1,
"count": 2,
"doubleBonds": {
"numDoubleBonds": 0,
"doubleBondPositions": {}
},
"functionalGroups": {},
"ndoubleBonds": 0
}
]
},
"numCarbon": 18,
"lipidFaBondType": "LCB_EXCEPTION",
"ndoubleBonds": 1
},
{
"name": "FA1",
"position": 2,
"count": 1,
"doubleBonds": {
"numDoubleBonds": 1,
"doubleBondPositions": {
"6": "Z"
}
},
"functionalGroups": {},
"numCarbon": 16,
"lipidFaBondType": "ESTER",
"ndoubleBonds": 1
}
],
"lipidLevel": "SN_POSITION",
"lipidString": "Cer 18:1;O2/16:1",
"extendedClass": "Cer"
},
"adduct": null,
"className": "Cer",
"lipidLevel": "SN_POSITION",
"extendedClass": "Cer",
"lipidString": "Cer 18:1;O2/16:1",
"mass": 535.4964451650001,
"sumFormula": "C34H65NO3"
},
"functionalGroupCounts": {
"OH": 2
},
"normalizedName": "Cer 18:1;O2/16:1",
"lipidMapsCategory": "SP",
"lipidMapsClass": "SP02",
"mass": 535.4964451650001,
"sumFormula": "C34H65NO3",
"lipidMapsReferences": [],
"swissLipidsReferences": []
}
],
"totalReceived": 1,
"totalParsed": 1,
"failedToParse": 0
}
The response part also reports the normalized name (goslinName), as
well as classification information using the LIPID MAPS® category and
class associated to the parsed lipid.
...
"normalizedName": "Cer 18:1;O2/16:1",
"lipidMapsCategory": "SP",
"lipidMapsClass": "SP02",
"mass": 535.4964451650001,
"sumFormula": "C34H65NO3",
...
The response also reports information on the fatty acyls detected in the lipid name. In this case, a Ceramide with a long-chain base and an additional fatty acyl has been detected. The name given here as an example was classified on SN_position level, since the lcb contains one double bond, but without positional E/Z information. The fatty acyl FA1 at the sn2 position does report E/Z information for its double bond.
...
"faList": [
{
"name": "LCB",
"position": 1,
"count": 1,
"doubleBonds": {
"numDoubleBonds": 1,
"doubleBondPositions": {}
},
"functionalGroups": {
"OH": [
{
"name": "OH",
"position": -1,
"count": 2,
"doubleBonds": {
"numDoubleBonds": 0,
"doubleBondPositions": {}
},
"functionalGroups": {},
"ndoubleBonds": 0
}
]
},
"numCarbon": 18,
"lipidFaBondType": "LCB_EXCEPTION",
"ndoubleBonds": 1
},
{
"name": "FA1",
"position": 2,
"count": 1,
"doubleBonds": {
"numDoubleBonds": 1,
"doubleBondPositions": {
"6": "Z"
}
},
"functionalGroups": {},
"numCarbon": 16,
"lipidFaBondType": "ESTER",
"ndoubleBonds": 1
}
],
...
Finally, the response reports the total number lipid names received, the total number parsed and the total number of parsing failures.
...
"totalReceived": 1,
"totalParsed": 1,
"failedToParse": 0
...
Lipid Classes supported by Goslin and the web application
| Lipid Category | LIPID MAPS Class Name | Allowed #s of FAs | Maximum # of FAs | Sum Formula (Head Group) | Mass (Head Group) | Synonyms |
|---|---|---|---|---|---|---|
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C20H34O4 | 338.2457 | ["11,12-DHET", 11,12-DiHETrE] |
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C20H34O4 | 338.2457 | ["14,15-DHET", 14,15-DiHETrE] |
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C20H32O4 | 336.2301 | ["5,12-DiHETE"] |
| FA | Lipoxins [FA0304] | 0 | 0 | C20H32O5 | 352.2250 | ["5,6,15-LXA4"] |
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C20H32O4 | 336.2301 | ["5,6-DiHETE"] |
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C20H34O4 | 338.2457 | ["8,9-DHET", 8,9-DiHETrE] |
| FA | Other Docosanoids [FA0400] | 0 | 0 | C22H32O3 | 344.2351 | [10-HDoHE] |
| FA | Epoxyeicosatrienoic acids [FA0308] | 0 | 0 | C20H32O3 | 320.2351 | [11(12)-EET, 11,12-EpETrE] |
| FA | Other Docosanoids [FA0400] | 0 | 0 | C22H32O3 | 344.2351 | [11-HDoHE] |
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C20H32O3 | 320.2351 | [11-HETE] |
| FA | Other Octadecanoids [FA0200] | 0 | 0 | C18H32O3 | 296.2351 | [12(13)-EpOME] |
| FA | Hydroxy/hydroperoxyeicosapentaenoic acids [FA0307] | 0 | 0 | C20H30O3 | 318.2195 | [12-HEPE] |
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C20H32O3 | 320.2351 | [12-HETE] |
| FA | Hydroxy/hydroperoxyeicosatrienoic acids [FA0305] | 0 | 0 | C17H28O3 | 280.2038 | [12-HHTrE] |
| FA | Fatty acids and conjugates [FA01] | 0 | 0 | C20H30O3 | 318.2195 | [12-OxoETE] |
| FA | Other Octadecanoids [FA0200] | 0 | 0 | C18H32O3 | 296.2351 | [13-HODE, 13R-HODE] |
| FA | Other Octadecanoids [FA0200] | 0 | 0 | C18H30O3 | 294.2195 | [13-HOTrE] |
| FA | Epoxyeicosatrienoic acids [FA0308] | 0 | 0 | C20H32O3 | 320.2351 | [14(15)-EET, 14,15-EpETrE] |
| FA | Other Eicosanoids [FA0300] | 0 | 0 | C20H30O3 | 318.2195 | [14(15)-EpETE] |
| FA | Hydroxy/hydroperoxyeicosapentaenoic acids [FA0307] | 0 | 0 | C20H30O3 | 318.2195 | [15-HEPE] |
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C20H32O3 | 320.2351 | [15-HETE] |
| FA | Prostaglandins [FA0301] | 0 | 0 | C20H28O3 | 316.2038 | [15d-PGJ2] |
| FA | Other Docosanoids [FA0400] | 0 | 0 | C22H32O3 | 344.2351 | [16-HDoHE] |
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C20H32O3 | 320.2351 | [16-HETE] |
| FA | Hydroxy/hydroperoxyeicosapentaenoic acids [FA0307] | 0 | 0 | C20H30O3 | 318.2195 | [18-HEPE] |
| FA | Epoxyeicosatrienoic acids [FA0308] | 0 | 0 | C20H32O3 | 320.2351 | [5(6)-EET, 5,6-EpETrE] |
| FA | Hydroxy/hydroperoxyeicosapentaenoic acids [FA0307] | 0 | 0 | C20H30O3 | 318.2195 | [5-HEPE] |
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C20H32O3 | 320.2351 | [5-HETE] |
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C20H32O4 | 336.2301 | [5-HpETE] |
| FA | Fatty acids and conjugates [FA01] | 0 | 0 | C20H30O3 | 318.2195 | [5-OxoETE] |
| FA | Epoxyeicosatrienoic acids [FA0308] | 0 | 0 | C20H32O3 | 320.2351 | [8(9)-EET, 8,9-EpETrE] |
| FA | Other Docosanoids [FA0400] | 0 | 0 | C22H32O3 | 344.2351 | [8-HDoHE] |
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C20H32O3 | 320.2351 | [8-HETE] |
| FA | Other Octadecanoids [FA0200] | 0 | 0 | C18H32O3 | 296.2351 | [9(10)-EpOME] |
| FA | Hydroxy/hydroperoxyeicosapentaenoic acids [FA0307] | 0 | 0 | C20H30O3 | 318.2195 | [9-HEPE] |
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C20H32O3 | 320.2351 | [9-HETE] |
| FA | Other Octadecanoids [FA0200] | 0 | 0 | C18H32O3 | 296.2351 | [9-HODE] |
| FA | Other Octadecanoids [FA0200] | 0 | 0 | C18H30O3 | 294.2195 | [9-HOTrE] |
| FA | Unsaturated fatty acids [FA0103] | 0 | 0 | C20H32O2 | 304.2402 | [AA, Arachidonic acid, Arachidonic Acid] |
| FA | Fatty acyl carnitines [FA0707] | 1 | 1 | C7H14NO3 | 160.0974 | [CAR] |
| FA | Fatty acyl CoAs [FA0705] | 1 | 1 | C21H35N7O16P3S | 766.1074 | [CoA] |
| FA | Unsaturated fatty acids [FA0103] | 0 | 0 | C22H32O2 | 328.2402 | [DHA, Docosapentaenoic acid] |
| FA | Unsaturated fatty acids [FA0103] | 0 | 0 | C20H30O2 | 302.2246 | [EPA, Eicosapentaenoic acid] |
| FA | Fatty acids and conjugates [FA01] | 1 | 1 | HO | 17.0027 | [FA, fatty acid] |
| FA | Wax monoesters [FA0701] | 1 | 1 | HO | 17.0027 | [FAHFA] |
| FA | Fatty aldehydes [FA06] | 1 | 1 | H | 1.0078 | [FAL] |
| FA | Fatty alcohols [FA05] | 1 | 1 | H3 | 3.0235 | [FOH, fatty alcohol] |
| FA | Glycerophosphoethanolamine | 1 | 1 | C5H13NO6P | 214.0480 | [GP-NAE] |
| FA | Hydrocarbons [FA11] | 1 | 1 | H | 1.0078 | [HC] |
| FA | Leukotrienes [FA0302] | 0 | 0 | C20H32O4 | 336.2301 | [LTB4] |
| FA | Eicosanoid derivatives | 0 | 0 | C30H47N3O9S | 625.3033 | [LTC4] |
| FA | Leukotrienes [FA0302] | 0 | 0 | C25H40N2O6S | 496.2607 | [LTD4] |
| FA | Unsaturated fatty acids [FA0103] | 0 | 0 | C18H32O2 | 280.2402 | [Linoleic acid] |
| FA | Maresins [FA0405] | 0 | 0 | C22H32O4 | 360.2301 | [Maresin 1] |
| FA | Fatty amides | 2 | 2 | HNO | 31.0058 | [NA] |
| FA | Fatty amides | 1 | 1 | C2H6NO | 60.0449 | [NAE] |
| FA | N-acyl amines [FA0802] | 1 | 1 | C2H6NO3S | 124.0068 | [NAT] |
| FA | Prostaglandins [FA0301] | 0 | 0 | C20H30O4 | 334.2144 | [PGB2] |
| FA | Prostaglandins [FA0301] | 0 | 0 | C20H32O5 | 352.2250 | [PGD2] |
| FA | Prostaglandins [FA0301] | 0 | 0 | C20H32O5 | 352.2250 | [PGE2] |
| FA | Prostaglandins [FA0301] | 0 | 0 | C20H34O5 | 354.2406 | [PGF2alpha] |
| FA | Prostaglandins [FA0301] | 0 | 0 | C20H32O5 | 352.2250 | [PGI2] |
| FA | Straight chain fatty acids [FA0101] | 0 | 0 | C16H32O2 | 256.2402 | [Palmitic acid] |
| FA | Resolvin Ds [FA0403] | 0 | 0 | C22H32O5 | 376.2250 | [Resolvin D1] |
| FA | Resolvin Ds [FA0403] | 0 | 0 | C22H32O5 | 376.2250 | [Resolvin D2] |
| FA | Resolvin Ds [FA0403] | 0 | 0 | C22H32O5 | 376.2250 | [Resolvin D3] |
| FA | Resolvin Ds [FA0403] | 0 | 0 | C22H32O4 | 360.2301 | [Resolvin D5] |
| FA | Thromboxanes [FA0303] | 0 | 0 | C20H36O6 | 372.2512 | [TXB1] |
| FA | Thromboxanes [FA0303] | 0 | 0 | C20H34O6 | 370.2355 | [TXB2] |
| FA | Thromboxanes [FA0303] | 0 | 0 | C20H32O6 | 368.2199 | [TXB3] |
| FA | Wax diesters [FA0702] | 2 | 2 | H2 | 2.0157 | [WD] |
| FA | Fatty esters [FA07] | 2 | 2 | O | 15.9949 | [WE] |
| FA | Fatty acids and conjugates [FA01] | 0 | 0 | N.D. | [alpha-LA, ALA] | |
| FA | Hydroxy/hydroperoxyeicosatetraenoic acids [FA0306] | 0 | 0 | C16H26O3 | 266.1882 | [tetranor-12-HETE] |
| GL | Diacylglycerols [GL0201] | 2 | 3 | C3H5O3 | 89.0239 | [DG, DAG] |
| GL | Other Glycerolipids [GL00] | 2 | 2 | C10H19NO6 | 249.1212 | [DGCC] |
| GL | Glycosyldiradylglycerols [GL05] | 2 | 2 | C15H26O13 | 414.1373 | [DGDG] |
| GL | Glycosyldiradylglycerols [GL05] | 1 | 2 | C15H26O13 | 414.1373 | [DGMG] |
| GL | Dihexosyldiacylglycerol | 2 | 2 | C15H26O13 | 414.1373 | [DHDG] |
| GL | Glycosyldiradylglycerols [GL05] | 2 | 2 | N.D. | [Glc-DG] | |
| GL | Monoacylglycerols [GL0101] | 1 | 3 | C3H5O3 | 89.0239 | [MG, MAG] |
| GL | Glycosyldiacylglycerols [GL0501] | 2 | 2 | C9H16O8 | 252.0845 | [MGDG] |
| GL | Glycosyldiacylglycerols [GL0501] | 1 | 2 | C9H16O8 | 252.0845 | [MGMG] |
| GL | Monohexosyldiacylglycerol | 2 | 2 | C9H16O8 | 252.0845 | [MHDG] |
| GL | Glycosylalkylacylglycerols [GL0502] | 2 | 2 | C11H22NO11P | 375.0930 | [PE-GlcDG] |
| GL | Glycosylalkylacylglycerols [GL0502] | 2 | 2 | C9H16O11S | 332.0413 | [SMGDG, Seminolipid] |
| GL | Glycosyldiacylglycerols [GL0501] | 2 | 2 | C9H16O10S | 316.0464 | [SQDG] |
| GL | Glycosylmonoacylglycerols [GL0401] | 1 | 2 | C9H16O10S | 316.0464 | [SQMG] |
| GL | Triacylglycerols [GL0301] | 3 | 3 | C3H5O3 | 89.0239 | [TG, TAG] |
| GL | Estolides [GL0305] | 3 | 3 | C3H5O3 | 89.0239 | [TG-EST, TG EST, TAG EST, TAG-EST] |
| GP | Diacylglycerophosphoinositol bisphosphates [GP0801] | 2 | 2 | C9H19O17P3 | 491.9835 | ["PIP2(3',4')", PIP2[3,4], PIP2[3',4']] |
| GP | Diacylglycerophosphoinositol bisphosphates [GP0801] | 2 | 2 | C9H19O17P3 | 491.9835 | ["PIP2(3',5')", PIP2[3,5], PIP2[3',5']] |
| GP | Diacylglycerophosphoinositol bisphosphates [GP0801] | 2 | 2 | C9H19O17P3 | 491.9835 | ["PIP2(4',5')", PIP2[4,5], PIP2[4',5']] |
| GP | Diacylglycerophosphoinositol trisphosphates [GP0901] | 2 | 2 | C9H20O20P4 | 571.9498 | ["PIP3(3',4',5')", PIP3[3,4,5], PIP3[3',4',5']] |
| GP | Glycosylglycerophospholipids [GP14] | 2 | 2 | C11H19O12P | 374.0614 | [6-Ac-Glc-GP] |
| GP | Monoacylglycerophosphomonoradylglycerols [GP0410] | 2 | 4 | C6H11O8P | 242.0192 | [BMP, LBPA] |
| GP | CDP-diacylglycerols [GP1301] | 2 | 2 | C12H19N3O13P2 | 475.0393 | [CDPDAG, CDPDG, CDP-DG, CDP-DAG] |
| GP | Glycerophosphoglycerophosphoglycerols [GP12] | 4 | 4 | C9H18O13P2 | 396.0223 | [CL] |
| GP | Glycerophosphoinositolglycans [GP15] | 1 | 1 | C3H6O5P | 152.9953 | [CPA, cLPA, CLPA] |
| GP | Glycerophosphoglycerophosphoglycerols [GP12] | 2 | 4 | C9H18O13P2 | 396.0223 | [DLCL] |
| GP | Dimethylphosphatidylethanolamine | 2 | 2 | C7H16NO6P | 241.0715 | [DMPE] |
| GP | Diacylglycosylglycerophospholipids [GP1401] | 2 | 2 | C9H17O11P | 332.0508 | [Glc-GP] |
| GP | Lyso-CDP-diacylglycerol | 1 | 2 | N.D. | [LCDPDAG] | |
| GP | Glycerophosphoglycerophosphoglycerols [GP12] | 3 | 4 | C9H18O13P2 | 396.0223 | [LCL, MLCL] |
| GP | Lysodimethylphosphatidylethanolamine | 1 | 2 | N.D. | [LDMPE] | |
| GP | Lysomonomethylphosphatidylethanolamine | 1 | 2 | C6H15NO6P | 228.0637 | [LMMPE] |
| GP | Monoacylglycerophosphates [GP1005] | 1 | 2 | C3H7O6P | 169.9980 | [LPA, LysoPA, lysoPA] |
| GP | Monoacylglycerophosphocholines [GP0105] | 1 | 2 | C8H18NO6P | 255.0872 | [LPC, LysoPC, lysoPC] |
| GP | Monoacylglycerophosphoethanolamines [GP0205] | 1 | 2 | C5H12NO6P | 213.0402 | [LPE, LysoPE, lysoPE] |
| GP | N-acyl/alkyl PE | 1 | 2 | C5H11NO6P | 212.0324 | [LPE-N, LPE-N(Alk), LPE-N(FA)] |
| GP | Monoacylglycerophosphoglycerols [GP0405] | 1 | 2 | C6H13O8P | 244.0348 | [LPG, LysoPG, lysoPG] |
| GP | Monoacylglycerophosphoinositols [GP0605] | 1 | 2 | C9H17O11P | 332.0508 | [LPI, LysoPI, lysoPI] |
| GP | Monoacylglycerophosphoinositolglycans [GP1504] | 1 | 2 | C15H27O16P | 494.1037 | [LPIM1] |
| GP | Monoacylglycerophosphoinositolglycans [GP1504] | 1 | 2 | C21H37O21P | 656.1565 | [LPIM2] |
| GP | Monoacylglycerophosphoinositolglycans [GP1504] | 1 | 2 | C27H47O26P | 818.2093 | [LPIM3] |
| GP | Monoacylglycerophosphoinositolglycans [GP1504] | 1 | 2 | C34H59O30P | 978.2829 | [LPIM4] |
| GP | Monoacylglycerophosphoinositolglycans [GP1504] | 1 | 2 | C39H67O36P | 1142.3150 | [LPIM5] |
| GP | Monoacylglycerophosphoinositolglycans [GP1504] | 1 | 2 | C45H77O41P | 1304.3678 | [LPIM6] |
| GP | Lysophosphatidylinositol- mannosideinositolphosphate | 1 | 2 | N.D. | [LPIMIP] | |
| GP | Lysophosphatidylinositol-glucosamine | 1 | 2 | N.D. | [LPIN] | |
| GP | Monoacylglycerophosphoserines [GP0305] | 1 | 2 | C6H12NO8P | 257.0301 | [LPS, LysoPS, lysoPS] |
| GP | Monomethylphosphatidylethanolamine | 2 | 2 | C6H14NO6P | 227.0559 | [MMPE] |
| GP | Diacylglycerophosphoethanolamines [GP0201] | 3 | 3 | C5H11NO6P | 212.0324 | [NAPE] |
| GP | Glycerophosphates [GP10] | 2 | 2 | C3H7O6P | 169.9980 | [PA, GPA, PlsA] |
| GP | Glycerophosphocholines [GP01] | 2 | 2 | C8H18NO6P | 255.0872 | [PC, GPC, GPCho, PlsCho] |
| GP | Glycerophosphoethanolamines [GP02] | 2 | 2 | C5H12NO6P | 213.0402 | [PE, GPE, GPEtn, PlsEtn] |
| GP | N-acyl/alkyl PE | 2 | 2 | C5H11NO6P | 212.0324 | [PE-N, PE-N(Alk), PE-N(FA)] |
| GP | Diacylglycerophosphoethanolamines [GP0201] | 2 | 2 | C6H14NO6P | 227.0559 | [PE-NMe] |
| GP | Diacylglycerophosphoethanolamines [GP0201] | 2 | 2 | C7H16NO6P | 241.0715 | [PE-NMe2] |
| GP | Glycerophosphoethanolamines [GP02] | 2 | 2 | C5H11O6P | 198.0293 | [PEt, PEtOH] |
| GP | Glycerophosphoglycerols [GP04] | 2 | 2 | C6H13O8P | 244.0348 | [PG, GPG, GPGro, PlsGro] |
| GP | Glycerophosphoglycerophosphates [GP05] | 2 | 2 | C6H14O11P2 | 324.0011 | [PGP] |
| GP | Glycerophosphoinositols [GP06] | 2 | 2 | C9H17O11P | 332.0508 | [PI, GPI, GPIns, PlsIns] |
| GP | Glycerophosphoinositolglycans [GP15] | 2 | 2 | C15H27O16P | 494.1037 | [PIM1] |
| GP | Glycerophosphoinositolglycans [GP15] | 2 | 2 | C21H37O21P | 656.1565 | [PIM2] |
| GP | Glycerophosphoinositolglycans [GP15] | 2 | 2 | C27H47O26P | 818.2093 | [PIM3] |
| GP | Glycerophosphoinositolglycans [GP15] | 2 | 2 | C33H57O31P | 980.2621 | [PIM4] |
| GP | Glycerophosphoinositolglycans [GP15] | 2 | 2 | C39H67O36P | 1142.3150 | [PIM5] |
| GP | Glycerophosphoinositolglycans [GP15] | 2 | 2 | C45H77O41P | 1304.3678 | [PIM6] |
| GP | Phosphatidylinositol mannoside inositol phosphate | 2 | 2 | N.D. | [PIMIP] | |
| GP | Diacylglycerophosphoinositol monophosphates [GP0701] | 2 | 2 | C9H18O14P2 | 412.0172 | [PIP] |
| GP | Diacylglycerophosphoinositol monophosphates [GP0701] | 2 | 2 | C9H18O14P2 | 412.0172 | [PIP(3'), PIP[3], PIP[3']] |
| GP | Diacylglycerophosphoinositol monophosphates [GP0701] | 2 | 2 | C9H18O14P2 | 412.0172 | [PIP(4'), PIP[4], PIP[4']] |
| GP | Diacylglycerophosphoinositol monophosphates [GP0701] | 2 | 2 | C9H18O14P2 | 412.0172 | [PIP(5'), PIP[5], PIP[5']] |
| GP | Diacylglycerophosphoinositol bisphosphates [GP0801] | 2 | 2 | C9H19O17P3 | 491.9835 | [PIP2] |
| GP | Diacylglycerophosphoinositol trisphosphates [GP0901] | 2 | 2 | C9H20O20P4 | 571.9498 | [PIP3] |
| GP | Glycerophosphoethanolamines [GP02] | 2 | 2 | C4H9O6P | 184.0137 | [PMeOH] |
| GP | Diacylglyceropyrophosphates [GP1101] | 2 | 2 | C3H8O9P2 | 249.9644 | [PPA] |
| GP | Glycerophosphoserines [GP03] | 2 | 2 | C6H12NO8P | 257.0301 | [PS, GPS, GPSer, PlsSer] |
| GP | N-acyl/alkyl PS | 2 | 2 | N.D. | [PS-N, PS-N(Alk), PS-N(FA)] | |
| GP | Diacylglycerophosphoserines [GP0301] | 3 | 3 | C6H11NO8P | 256.0222 | [PS-NAc] |
| GP | Other Glycerophospholipids [GP00] | 2 | 2 | C7H14NO8P | 271.0457 | [PT] |
| GP | Glycerophosphonocholines [GP16] | 2 | 2 | C8H18NO5P | 239.0923 | [PnC] |
| GP | Glycerophosphoinositolglycans [GP15] | 2 | 2 | C5H12NO5P | 197.0453 | [PnE] |
| GP | Diacylglycerophosphomonoradylglycerols [GP0409] | 3 | 4 | C6H11O8P | 242.0192 | [SLBPA] |
| NO_CATEGORY | 0 | 0 | N.D. | [UNDEFINED] | ||
| PK | Anacardic acids and derivatives [PK1504] | 2 | 2 | N.D. | [ANACARD] | |
| PK | Alkyl catechols and derivatives [PK1502] | 2 | 2 | N.D. | [CATECHOL] | |
| PK | Alkyl phenols and derivatives [PK1501] | 2 | 2 | N.D. | [PHENOL] | |
| PK | Alkyl resorcinols and derivatives [PK1503] | 2 | 2 | N.D. | [RESORCINOL] | |
| SL | Acyltrehaloses [SL03] | 2 | 2 | C20H36O14S | 532.1826 | [AC2SGL] |
| SL | Acyltrehaloses [SL03] | 2 | 2 | C15H26O11 | 382.1475 | [DAT] |
| SL | Acyltrehaloses [SL03] | 4 | 4 | N.D. | [PAT16] | |
| SL | Acyltrehaloses [SL03] | 4 | 4 | N.D. | [PAT18] | |
| SP | Glycosphingolipids | 2 | 2 | C12H20O13S | 404.0625 | [(3'-sulfo)LacCer] |
| SP | Glycosphingolipids | 2 | 2 | C24H41O19 | 633.2242 | [(Fuc)iGb3Cer] |
| SP | Acylceramides [SP0204] | 2 | 2 | C22H43O | 323.3314 | [1-O-behenoyl-Cer] |
| SP | Acylceramides [SP0204] | 2 | 2 | C28H55O | 407.4253 | [1-O-carboceroyl-Cer] |
| SP | Acylceramides [SP0204] | 2 | 2 | C26H51O | 379.3940 | [1-O-cerotoyl-Cer] |
| SP | Acylceramides [SP0204] | 2 | 2 | C20H39O | 295.3001 | [1-O-eicosanoyl-Cer] |
| SP | Acylceramides [SP0204] | 2 | 2 | C24H47O | 351.3627 | [1-O-lignoceroyl-Cer] |
| SP | Acylceramides [SP0204] | 2 | 2 | C14H27O | 211.2062 | [1-O-myristoyl-Cer] |
| SP | Acylceramides [SP0204] | 2 | 2 | C16H31O | 239.2375 | [1-O-palmitoyl-Cer] |
| SP | Acylceramides [SP0204] | 2 | 2 | C18H35O | 267.2688 | [1-O-stearoyl-Cer] |
| SP | Acylceramides [SP0204] | 2 | 2 | C70H59O | 915.4566 | [1-O-tricosanoyl-Cer] |
| SP | Ceramide [SP0204] | 2 | 2 | N.D. | [ACer] | |
| SP | Globoside | 2 | 2 | C50H78N3O37 | 1312.4314 | [Ac-O-9-GD1a] |
| SP | Globoside | 2 | 2 | C61H94N4O45 | 1602.5190 | [Ac-O-9-GT1b] |
| SP | Globoside | 2 | 2 | C47H71N3O35 | 1237.3868 | [Ac-O-9-GT3] |
| SP | Glycosphingolipids | 2 | 2 | C48H80N3O35 | 1258.4572 | [Branched-Forssman] |
| SP | Ceramides [SP02] | 2 | 2 | H | 1.0078 | [Cer, Ceramide] |
| SP | Ceramide 1-phosphates [SP0205] | 2 | 2 | H2O3P | 80.9742 | [CerP, C1P] |
| SP | Glycosphingolipids | 2 | 2 | C54H86N3O41 | 1432.4737 | [DSGG] |
| SP | Ceramide phosphoethanolamines [SP0302] | 2 | 2 | C2H7NO3P | 124.0164 | [EPC, PE-Cer, CerPE] |
| SP | Simple Glc series [SP0501] | 2 | 2 | C16H21O10 | 373.1135 | [FMC-5] |
| SP | Neutral glycosphingolipids [SP05] | 2 | 2 | C9H18O11P | 333.0587 | [FMC-6] |
| SP | Glycosphingolipids | 2 | 2 | C34H57N2O25 | 893.3250 | [Forssman] |
| SP | Acidic glycosphingolipids [SP06] | 2 | 2 | C49H80N2O37 | 1288.4440 | [Fuc(Gal)-GM1] |
| SP | Glycosphingolipids | 2 | 2 | C44H74NO34 | 1160.4092 | [Fuc(Gal)Gal-iGb4Cer] |
| SP | Glycosphingolipids | 2 | 2 | C54H90N3O39 | 1404.5151 | [Fuc-Branched-Forssman] |
| SP | Globoside | 2 | 2 | C32H54NO24 | 836.3036 | [Fuc-GA1] |
| SP | Globoside | 2 | 2 | C54H86N3O40 | 1416.4788 | [Fuc-GD1b] |
| SP | Globoside | 2 | 2 | C43H70N2O32 | 1126.3912 | [Fuc-GM1] |
| SP | Globoside | 2 | 2 | C43H70N2O33 | 1142.3861 | [Fuc-GM1(NeuGc)] |
| SP | Glycosphingolipids | 2 | 2 | C24H41O19 | 633.2242 | [Fuc-iGb3Cer] |
| SP | Glycosphingolipids | 2 | 2 | C30H51O24 | 795.2770 | [FucGalGb3Cer] |
| SP | Glycosphingolipids | 2 | 2 | C26H44NO20 | 690.2457 | [GA1, Ga1] |
| SP | Glycosphingolipids | 2 | 2 | C20H34NO15 | 528.1928 | [GA2, Ga2] |
| SP | Neutral glycosphingolipids [SP05] | 2 | 2 | C18H31O15 | 487.1663 | [GB3, Gb3, GB3Cer, Gb3Cer] |
| SP | Neutral glycosphingolipids [SP05] | 2 | 2 | C26H44NO20 | 690.2457 | [GB4, Gb4, Tetrahexosylceramide, Gb4Cer, GB4Cer] |
| SP | Glycosphingolipids | 2 | 2 | C48H76N3O36 | 1270.4209 | [GD1, Gd1] |
| SP | Ganglioside | 2 | 2 | C48H76N3O36 | 1270.4209 | [GD1a, Gd1a] |
| SP | Ganglioside | 2 | 2 | C48H76N3O36 | 1270.4209 | [GD1a alpha] |
| SP | Ganglioside | 2 | 2 | C48H76N3O36 | 1270.4209 | [GD1a(NeuAc/NeuGc)] |
| SP | Ganglioside | 2 | 2 | C48H76N3O37 | 1286.4158 | [GD1a(NeuGc/NeuAc)] |
| SP | Ganglioside | 2 | 2 | C48H76N3O38 | 1302.4107 | [GD1a(NeuGc/NeuGc)] |
| SP | Ganglioside | 2 | 2 | C48H76N3O36 | 1270.4209 | [GD1b, Gd1b] |
| SP | Ganglioside | 2 | 2 | C48H76N3O36 | 1270.4209 | [GD1c, Gd1c] |
| SP | Ganglioside | 2 | 2 | C48H76N3O38 | 1302.4107 | [GD1c(NeuGc/NeuGc)] |
| SP | Glycosphingolipids | 2 | 2 | C42H66N3O31 | 1108.3680 | [GD2, Gd2] |
| SP | Glycosphingolipids | 2 | 2 | C34H55N2O26 | 907.3043 | [GD3, Gd3] |
| SP | Glycosphingolipids | 2 | 2 | C37H60N2O28 | 980.3333 | [GM1, Gm1] |
| SP | Ganglioside | 2 | 2 | C37H60N2O28 | 980.3333 | [GM1 alpha] |
| SP | Ganglioside | 2 | 2 | C37H60N2O28 | 980.3333 | [GM1(NeuGc)] |
| SP | Ganglioside | 2 | 2 | C37H60N2O28 | 980.3333 | [GM1b] |
| SP | Ganglioside | 2 | 2 | C37H60N2O29 | 996.3282 | [GM1b(NeuGc)] |
| SP | Glycosphingolipids | 2 | 2 | C31H50N2O23 | 818.2804 | [GM2, Gm2] |
| SP | Ganglioside | 2 | 2 | C31H50N2O24 | 834.2754 | [GM2(NeuGc)] |
| SP | Glycosphingolipids | 2 | 2 | C23H38NO18 | 616.2089 | [GM3, Gm3] |
| SP | Gangliosides [SP0601] | 2 | 2 | C17H28NO13 | 454.1561 | [GM4, Gm4] |
| SP | Glycosphingolipids | 2 | 2 | C81H124N6O60 | 2140.6836 | [GP1, Gp1] |
| SP | Ganglioside | 2 | 2 | C81H124N6O60 | 2140.6836 | [GP1c] |
| SP | Ganglioside | 2 | 2 | C81H124N6O60 | 2140.6836 | [GP1c alpha] |
| SP | Glycosphingolipids | 2 | 2 | C70H108N5O52 | 1850.5960 | [GQ1, Gq1] |
| SP | Ganglioside | 2 | 2 | C70H108N5O52 | 1850.5960 | [GQ1b] |
| SP | Ganglioside | 2 | 2 | C70H108N5O52 | 1850.5960 | [GQ1b alpha] |
| SP | Ganglioside | 2 | 2 | C70H108N5O52 | 1850.5960 | [GQ1c] |
| SP | Glycosphingolipids | 2 | 2 | C59H92N4O44 | 1560.5084 | [GT1, Gt1] |
| SP | Ganglioside | 2 | 2 | C59H92N4O44 | 1560.5084 | [GT1a] |
| SP | Ganglioside | 2 | 2 | C59H92N4O44 | 1560.5084 | [GT1a alpha] |
| SP | Ganglioside | 2 | 2 | C59H92N4O44 | 1560.5084 | [GT1b] |
| SP | Globoside | 2 | 2 | C59H92N4O44 | 1560.5084 | [GT1b alpha] |
| SP | Globoside | 2 | 2 | C59H92N4O45 | 1576.5034 | [GT1b alpha(NeuGc)] |
| SP | Ganglioside | 2 | 2 | C59H92N4O44 | 1560.5084 | [GT1c] |
| SP | Glycosphingolipids | 2 | 2 | C53H82N4O39 | 1398.4556 | [GT2, Gt2] |
| SP | Glycosphingolipids | 2 | 2 | C45H69N3O34 | 1195.3762 | [GT3, Gt3] |
| SP | Globoside | 2 | 2 | C38H64NO29 | 998.3564 | [Gal(Fuc)-GA1] |
| SP | Globoside | 2 | 2 | C60H96N3O45 | 1578.5316 | [Gal(Fuc)-GD1b] |
| SP | Globoside | 2 | 2 | C54H86N3O41 | 1432.4737 | [Gal-GD1b] |
| SP | Glycosphingolipids | 2 | 2 | C32H54NO25 | 852.2985 | [Gal-iGb4Cer] |
| SP | Neutral glycosphingolipids [SP05] | 2 | 2 | C6H11O5 | 163.0606 | [GalCer] |
| SP | Globoside | 2 | 2 | C60H96N3O46 | 1594.5265 | [GalGal-GD1b] |
| SP | Glycosphingolipids | 2 | 2 | C36H61O30 | 973.3248 | [GalGalGalGb3Cer] |
| SP | Glycosphingolipids | 2 | 2 | C30H51O25 | 811.2719 | [GalGalGb3Cer] |
| SP | Globoside | 2 | 2 | C51H83N3O39 | 1361.4604 | [GalGalNAc-GM1b(NeuGc)] |
| SP | Glycosphingolipids | 2 | 2 | C24H41O20 | 649.2191 | [GalGb3Cer] |
| SP | Glycosphingolipids | 2 | 2 | C32H54NO25 | 852.2985 | [GalGb4Cer] |
| SP | Glycosphingolipids | 2 | 2 | C46H77N2O35 | 1217.4307 | [GalGlcNAc-GalGb4Cer] |
| SP | Globoside | 2 | 2 | C56H89N4O41 | 1473.5002 | [GalNAc-GD1a] |
| SP | Globoside | 2 | 2 | C56H89N4O42 | 1489.4951 | [GalNAc-GD1a(NeuAc/NeuGc)] |
| SP | Globoside | 2 | 2 | C56H89N4O42 | 1489.4951 | [GalNAc-GD1a(NeuGc/NeuAc)] |
| SP | Globoside | 2 | 2 | C45H73N3O33 | 1183.4126 | [GalNAc-GM1] |
| SP | Globoside | 2 | 2 | C45H73N3O33 | 1183.4126 | [GalNAc-GM1b] |
| SP | Globoside | 2 | 2 | C45H73N3O34 | 1199.4075 | [GalNAc-GM1b(NeuGc)] |
| SP | Globoside | 2 | 2 | C46H77N2O34 | 1201.4358 | [GalNAcGal(Fuc)-GA1] |
| SP | Glycosphingolipids | 2 | 2 | C32H54NO25 | 852.2985 | [GalNAcGalGb3Cer] |
| SP | Neutral glycosphingolipids [SP05] | 2 | 2 | C6H11O5 | 163.0606 | [GlcCer] |
| SP | Glycosphingolipids | 2 | 2 | C40H67N2O30 | 1055.3779 | [GlcNAc-GalGb4Cer] |
| SP | Glycosphingolipids | 2 | 2 | C26H44NO20 | 690.2457 | [GlcNAcGb3Cer] |
| SP | Glycosphingolipids | 2 | 2 | C46H77N2O34 | 1201.4358 | [Globo-A] |
| SP | Glycosphingolipids | 2 | 2 | C44H74NO34 | 1160.4092 | [Globo-B] |
| SP | Glycosphingolipids | 2 | 2 | C38H64NO29 | 998.3564 | [Globo-H] |
| SP | Glycosphingolipids | 2 | 2 | C52H87N2O39 | 1363.4886 | [Globo-Lex-9] |
| SP | Neutral glycosphingolipids [SP05] | 2 | 2 | C12H21O10 | 325.1135 | [Hex2Cer, CDH, DHCer, DHCER] |
| SP | Neutral glycosphingolipids [SP05] | 2 | 2 | C18H31O15 | 487.1663 | [Hex3Cer] |
| SP | Neutral glycosphingolipids [SP05] | 2 | 2 | C6H11O5 | 163.0606 | [HexCer, Glucosylceramide, CMH, CMH-OH, MHCER, MHCer] |
| SP | Ceramide phosphoinositols [SP0303] | 2 | 2 | C6H12O8P | 243.0270 | [IPC, PI-Cer] |
| SP | Neutral glycosphingolipids [SP05] | 1 | 1 | C12H22O10 | 326.1213 | [LHex2Cer, LDHCer, LDHCER] |
| SP | Neutral glycosphingolipids [SP05] | 1 | 1 | C18H32O15 | 488.1741 | [LHex3Cer, LTHCer, LTHCER] |
| SP | Hexosylsphingosine | 1 | 1 | C6H12O5 | 164.0685 | [LHexCer, HexSph] |
| SP | 1 | 1 | C6H11O8P | 242.0192 | [LIPC] | |
| SP | Ceramides [SP02] | 1 | 1 | C5H14NO3P | 167.0711 | [LSM, SPC, LysoSM] |
| SP | Neutral glycosphingolipids [SP05] | 2 | 2 | C12H21O10 | 325.1135 | [LacCer] |
| SP | Globoside | 2 | 2 | C57H93N3O42 | 1491.5234 | [Lex-GM1] |
| SP | Phosphosphingolipids [SP03] | 2 | 2 | C18H33O21P2 | 647.0990 | [M(IP)2C, MIP2C] |
| SP | Phosphosphingolipids [SP03] | 2 | 2 | C12H22O13P | 405.0798 | [MIPC] |
| SP | Glycosphingolipids | 2 | 2 | C43H70N2O33 | 1142.3861 | [MSGG] |
| SP | Glycosphingolipids | 2 | 2 | C32H54NO25 | 852.2985 | [NOR1] |
| SP | Glycosphingolipids | 2 | 2 | C46H77N2O35 | 1217.4307 | [NOR2] |
| SP | Glycosphingolipids | 2 | 2 | C40H67N2O30 | 1055.3779 | [NORint] |
| SP | Glycosphingolipids | 2 | 2 | C54H86N3O41 | 1432.4737 | [NeuAc(alpha2-6)-MSGG] |
| SP | Glycosphingolipids | 2 | 2 | C54H86N3O41 | 1432.4737 | [NeuAc(alpha2-8)-MSGG] |
| SP | Glycosphingolipids | 2 | 2 | C43H70N2O33 | 1142.3861 | [NeuAcGal-iGb4Cer] |
| SP | Glycosphingolipids | 2 | 2 | C43H70N2O34 | 1158.3810 | [NeuGc-GalGb4Cer] |
| SP | Globoside | 2 | 2 | C62H99N4O48 | 1667.5429 | [NeuGc-LacNAc-GM1(NeuGc)] |
| SP | Glycosphingolipids | 2 | 2 | C54H86N3O43 | 1464.4635 | [NeuGcNeuGc-GalGb4Cer] |
| SP | Glycosphingolipids | 2 | 2 | C34H57N2O25 | 893.3250 | [Para-Forssman] |
| SP | Sulfoglycosphingolipids (sulfatides) [SP0602] | 2 | 2 | C26H42NO26S2 | 848.1436 | [SB1a] |
| SP | Sulfoglycosphingolipids (sulfatides) [SP0602] | 2 | 2 | C12H19O14S | 419.0496 | [SHex2Cer, SulfoHex2Cer] |
| SP | Sulfoglycosphingolipids (sulfatides) [SP0602] | 2 | 2 | C6H11O8S | 243.0175 | [SHexCer, SulfoHexCer, (3'-sulfo)Galbeta-Cer, (3'-sulfo)GalCer] |
| SP | Sulfonolipid | 2 | 2 | HO3S | 80.9646 | [SL] |
| SP | Ceramide phosphocholines (sphingomyelins) [SP0301] | 2 | 2 | C5H13NO3P | 166.0633 | [SM] |
| SP | Globoside | 2 | 2 | C26H43NO23S | 769.1947 | [SM1a] |
| SP | Globoside | 2 | 2 | C26H43NO23S | 769.1947 | [SM1b] |
| SP | Globoside | 2 | 2 | C37H59N2O32S | 1075.2772 | [SO3-GM1(NeuGc)] |
| SP | Glycosphingolipids | 2 | 2 | C34H56N2O28S | 972.2740 | [SO3-Gal-iGb4Cer] |
| SP | Glycosphingolipids | 2 | 2 | C32H53NO28S | 931.2475 | [SO3-GalGb4Cer] |
| SP | Glycosphingolipids | 2 | 2 | C26H43NO23S | 769.1947 | [SO3-Gb4Cer] |
| SP | Glycosphingolipids | 2 | 2 | C26H43NO23S | 769.1947 | [SO3-iGb4Cer] |
| SP | Sphingoid base homologs and variants [SP0104] | 1 | 1 | H2 | 2.0157 | [SPB, Sphingosine, So, Sphinganine, Sa, SPH, Sph, LCB, SPA] |
| SP | Sphingoid base 1-phosphates [SP0105] | 1 | 1 | H3O3P | 81.9820 | [SPBP, Sphingosine-1-phosphate, S1P, SPH-P, SIP, Sphinganine-1-phosphate, Sa1P, LCBP, SPA1P] |
| SP | Glycosphingolipids | 2 | 2 | C6H11O8S | 243.0175 | [SulfoGalCer] |
| SP | Glycosphingolipids | 2 | 2 | C34H57N2O25 | 893.3250 | [i-Forssman] |
| SP | Glycosphingolipids | 2 | 2 | C18H31O15 | 487.1663 | [iGb3Cer] |
| SP | Glycosphingolipids | 2 | 2 | C26H44NO20 | 690.2457 | [iGb4Cer] |
| ST | Monoradylglycosterols | 1 | 1 | H | 1.0078 | [ASG] |
| ST | Bile acids and derivatives [ST04] | 1 | 1 | H | 1.0078 | [BA] |
| ST | Sterol esters [ST0102] | 1 | 1 | C27H45O | 385.3470 | [SE 27:1, ChE, CE, Cholesteryl ester, Cholesterol ester, SE, Sterol ester, ChoE, CholE] |
| ST | Sterol esters [ST0102] | 1 | 1 | C27H43O | 383.3314 | [SE 27:2, Desmosterol ester] |
| ST | Sterol esters [ST0102] | 1 | 1 | C28H45O | 397.3470 | [SE 28:2, Ergostadienol ester] |
| ST | Sterol esters [ST0102] | 1 | 1 | C28H43O | 395.3314 | [SE 28:3, Ergosterol ester] |
| ST | Sterol esters [ST0102] | 1 | 1 | C29H49O | 413.3783 | [SE 29:1] |
| ST | Sterol esters [ST0102] | 1 | 1 | C29H47O | 411.3627 | [SE 29:2, Stigmasterol ester] |
| ST | Sterol esters [ST0102] | 1 | 1 | C30H49O | 425.3783 | [SE 30:2, Lanosterol ester] |
| ST | Sterylglycosides | 1 | 1 | H | 1.0078 | [SG] |
| ST | Sulfates [ST0502] | 0 | 0 | C27H48O4S | 468.3273 | [ST 27:0;O;S] |
| ST | Cholesterol and derivatives [ST0101] | 0 | 0 | C27H46O | 386.3549 | [ST 27:1;O, CH, FC, Cholesterol, Ch, ST, ST 27:1;1, Chol, cholesterol] |
| ST | Cholesterol and derivatives [ST0101] | 0 | 0 | C33H56O6 | 548.4077 | [ST 27:1;O;Hex, SG 27:1;O;Hex] |
| ST | Sulfates [ST0502] | 0 | 0 | C27H46O4S | 466.3117 | [ST 27:1;O;S, Cholesterol sulfate] |
| ST | Cholesterol and derivatives [ST0101] | 0 | 0 | C27H44O | 384.3392 | [ST 27:2;O, Desmosterol, ST 27:2;1] |
| ST | Cholesterol and derivatives [ST0101] | 0 | 0 | C28H48O | 400.3705 | [ST 28:1;O, Campesterol, ST 28:1;1] |
| ST | Ergosterols and C24-methyl derivatives [ST0103] | 0 | 0 | C34H58O6 | 562.4233 | [ST 28:1;O;Hex, SG 28:1;O;Hex] |
| ST | Sulfates [ST0502] | 0 | 0 | C28H48O4S | 480.3273 | [ST 28:1;O;S] |
| ST | Ergosterols and C24-methyl derivatives [ST0103] | 0 | 0 | C28H46O | 398.3549 | [ST 28:2;O, Ergostadienol, ST 28:2;1] |
| ST | Ergosterols and C24-methyl derivatives [ST0103] | 0 | 0 | C34H56O6 | 560.4077 | [ST 28:2;O;Hex, SG 28:2;O;Hex] |
| ST | Sulfates [ST0502] | 0 | 0 | C28H46O4S | 478.3117 | [ST 28:2;O;S, 24-methylene-cholesterol sulfate] |
| ST | Ergosterols and C24-methyl derivatives [ST0103] | 0 | 0 | C28H44O | 396.3392 | [ST 28:3;O, Ergosterol, ST 28:3;1] |
| ST | Cholesterol and derivatives [ST0101] | 0 | 0 | C29H50O | 414.3862 | [ST 29:1;O, Sitosterol, ST 29:1;1] |
| ST | Sulfates [ST0502] | 0 | 0 | C29H50O4S | 494.3430 | [ST 29:1;O;S] |
| ST | Stigmasterols and C24-ethyl derivatives [ST0104] | 0 | 0 | C29H48O | 412.3705 | [ST 29:2;O, Stigmasterol, ST 29:2;1] |
| ST | Cholesterol and derivatives [ST0101] | 0 | 0 | C30H50O | 426.3862 | [ST 30:2;O, Lanosterol, ST 30:2;1] |
| UNDEFINED | Undefined lipid class | 0 | 0 | N.D. | [UNDEFINED] |
Contributing
If you find that Goslin does not report a lipid name correctly, or is missing some information, please file a bug report or pull-request at our GitHub repository .
Please see the Goslin GitHub repository for the other programming language implementations and for the lipid-list.csv file, which contains information on which lipid head groups Goslin currently supports.
Using the Goslin implementations
The Goslin grammars are supported by parser implementations in multiple programming languages and use a similar object model. For details, please see the documentation at GitHub.
Citing Goslin
- Goslin 2.0 implements the recent lipid shorthand nomenclature for MS-derived lipid structures. Dominik Kopczynski, Nils Hoffmann, Bing Peng, Gerhard Liebisch, Friedrich Spener, Robert Ahrends. Preprint 2021
- Goslin - A Grammar of Succinct Lipid Nomenclature. Dominik Kopczynski, Nils Hoffmann, Bing Peng, Robert Ahrends. Analytical Chemistry 2020 92 (16), 10957-10960
Acknowledgements
In order to test lipid name parsing, sum formula calculation and cross-linking to reported lipids, the following resources were crucial during development and testing of Goslin and the Goslin web application:
Please see the Goslin GitHub repository for details on the database versions, dates and data that we used for development and testing.


